自实现核SVM用于分类任务 支持向量机是在分类与回归分析中分析数据的监督式学习模型与相关的学习算法。给定一组训练实例,每个训练实例被标记为属于两个类别中的一个或另一个,SVM训练算法创建一个将新的实例分配给两个类别之一的模型,使其成为非概率二元线性分类器。SVM模型是将实例表示为空间中的点,这样映射就使得单独类别的实例被尽可能宽的明显的间隔分开。然后,将新的实例映射到同一空间,并基于它们落在间隔的哪一侧来预测所属类别。
在这里,考虑基于核方法的支持向量机,并使用SMO算法去训练它,这是LIBSVM和sklearn中训练核SVM的方法,但在project1中我自行实现SMO算法求解器。
SMO算法 SMO(Sequential Minimal Optimization)是求解SVM问题的高效算法之一,libSVM采用的正是该算法。SMO算法其实是一种启发式算法:先选择两个变量 $α_i$ 和 $α_j$ ,然后固定其他参数,从而将问题转化成一个二变量的二次规划问题。求出能使目标最大的一对 $α_i$ 和$α_j$ 后,将它们固定,再选择两个变量,直到目标值收敛。
采用Chang, Chih-Chung, and Chih-Jen Lin. “LIBSVM: a library for support vector machines.” ACM transactions on intelligent systems and technology (TIST) 2.3 (2011): 1-27.中的实现方法。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 import numpy as npfrom functools import lru_cacheclass Solver :r'''SMO算法求解器,迭代求解下面的问题: .. math:: \min_{\pmb\alpha}\quad&\frac12\pmb\alpha^T\pmb Q\pmb\alpha+\pmb p^T\pmb\alpha\\ \text{s.t.}\quad&\pmb y^T\pmb\alpha=0\\ &0\leq\alpha_i\leq C,i=1,\cdots,l Parameters ---------- Q : numpy.ndarray 优化问题中的 :math:`\pmb Q` 矩阵; p : numpy.ndarray 优化问题中的 :math:`\pmb p` 向量; y : numpy.ndarray 优化问题中的 :math:`\pmb y` 向量; C : float 优化问题中的 :math:`C` 变量; tol : float, default=1e-5 变量选择的tolerance,默认为1e-5. ''' def __init__ (self, Q: np.ndarray, p: np.ndarray, y: np.ndarray, C: float , tol: float = 1e-5 ) -> None :0 ]assert problem_size == y.shape[0 ]if Q is not None :assert problem_size == Q.shape[0 ]assert problem_size == Q.shape[1 ]def working_set_select (self ):r'''工作集选择,这里采用一阶选择: .. math:: \pmb{I}_{up}(\pmb\alpha)&=\{t|\alpha_t<C,y_t=1\text{ or }\alpha_t>0,y_t=-1\}\\ \pmb{I}_{low}(\pmb\alpha)&=\{t|\alpha_t<C,y_t=-1\text{ or }\alpha_t>0,y_t=1\}\\ i&\in\arg\max_{t}\{-y_t\nabla_tf(\pmb\alpha)|t\in\pmb{I}_{up}(\pmb\alpha)\}\\ j&\in\arg\max_{t}\{-y_t\nabla_tf(\pmb\alpha)|t\in\pmb{I}_{low}(\pmb\alpha)\}\\ ''' 0 ),0 , self.y < 0 ),0 ),0 , self.y > 0 ),False try :except :True if find_fail or self.neg_y_grad[i] - self.neg_y_grad[j] < self.tol:return -1 , -1 return i, jdef update (self, i: int , j: int , func=None ):'''变量更新,在保证变量满足约束的条件下对两变量进行更新''' 2 * yi * yj * Qi[j]if quad_coef <= 0 :1e-12 if yi * yj == -1 :if diff > 0 :if (self.alpha[j] < 0 ):0 else :if (self.alpha[i] < 0 ):0 if diff > 0 :if (self.alpha[i] > self.C):else :if (self.alpha[j] > self.C):else :sum = self.alpha[i] + self.alpha[j]if sum > self.C:if self.alpha[i] > self.C:sum - self.Celse :if self.alpha[j] < 0 :0 sum if sum > self.C:if self.alpha[j] > self.C:sum - self.Celse :if self.alpha[i] < 0 :0 sum return delta_i, delta_jdef calculate_rho (self ) -> float :r'''计算偏置项 .. math:: \rho=\dfrac{\sum_{i:0<\alpha_i<C}y_i\nabla_if(\pmb\alpha)}{|\{i\vert0<\alpha_i<C\}|} 如果不存在满足条件的支持向量,那么 .. math:: -M(\pmb\alpha)&=\max\{y_i\nabla_if(\pmb\alpha)|\alpha_i=0,y_i=-1\text{ or }\alpha_i=C,y_i=1\}\\ -m(\pmb\alpha)&=\max\{y_i\nabla_if(\pmb\alpha)|\alpha_i=0,y_i=1\text{ or }\alpha_i=C,y_i=-1\}\\ \rho&=-\dfrac{M(\pmb\alpha)+m(\pmb\alpha)}{2} ''' 0 ,if sv.sum () > 0 :else :0 , self.y < 0 ),0 ),0 , self.y > 0 ),0 ),min () +max ()) / 2 return rhodef get_Q (self, i: int , func=None ):'''获取核矩阵的第i行/列,即 .. math:: [K(\pmb x_1, \pmb x_i),\cdots,K(\pmb x_l, \pmb x_i)] ''' return self.Q[i]
SVM接口的实现 为了让模型能够进行参数选择等功能,按照sklearn接口的模式编写SVM模型:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 from sklearn.base import BaseEstimatorfrom sklearn.metrics import accuracy_scorefrom solver import Solver, SolverWithCacheclass BiLinearSVC (BaseEstimator ):r'''二分类线性SVM,该类被多分类LinearSVC继承,所以不需要使用它。 通过求解对偶问题 .. math:: \min_{\pmb\alpha}\quad&\dfrac12\pmb\alpha^\top Q\pmb\alpha-\pmb{e}^\top\pmb{\alpha}\\ \text{s.t.}\quad& \pmb{y}^\top\pmb\alpha=0,\\ &0\leqslant\alpha_i\leqslant C,i=1,\cdots ,l 得到决策边界 .. math:: f(\pmb x)=\sum_{i=1}^ly_i\alpha_i\pmb x_i^T\pmb x-\rho Parameters ---------- C : float, default=1 SVM的正则化参数,默认为1; max_iter : int, default=1000 SMO算法迭代次数,默认1000; tol : float, default=1e-5 SMO算法的容忍度参数,默认1e-5; cache_size : int, default=256 lru缓存大小,默认256,如果为0则不使用缓存,计算Q矩阵然后求解. ''' def __init__ (self, C: float = 1. , max_iter: int = 1000 , tol: float = 1e-5 , cache_size: int = 256 ) -> None :super ().__init__()def fit (self, X: np.ndarray, y: np.ndarray ):'''训练模型 Parameters ---------- X : np.ndarray 训练集特征; y : np.array 训练集标签,建议0为负标签,1为正标签. ''' float )1 ] = -1 if self.cache_size == 0 :1 , 1 ) * y * np.matmul(X, X.T)else :def func (i ):return y * np.matmul(X, X[i]) * y[i]for n_iter in range (self.max_iter):if i < 0 :break else :print ("LinearSVC not coverage with {} iterations" .format (return selfdef decision_function (self, X: np.ndarray ) -> np.ndarray:'''决策函数,输出预测值''' return np.matmul(self.coef_[0 ], np.array(X).T) - self.coef_[-1 ]def predict (self, X: np.ndarray ) -> np.ndarray:'''预测函数,输出预测标签(0-1)''' return (self.decision_function(np.array(X)) >= 0 ).astype(int )def score (self, X: np.ndarray, y: np.ndarray ) -> float :'''评估函数,给定特征和标签,输出正确率''' return accuracy_score(y, self.predict(X))
模型训练 选用UCI数据集中的威斯康辛乳腺癌数据 作为实验数据集,首先查看数据分布等信息。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 import numpy as npfrom tqdm import tqdmfrom sklearn.preprocessing import LabelEncoderdef read_wdbc ():with open ("data/wdbc.data" , "r" ) as f:for line in tqdm(lines, desc='reading data' ):',' )[1 :]1 :])0 ])return (float ),print (X.shape, y.shape)
reading data: 100%|██████████████████████████████████████████████████████████████████████████| 569/569 [00:00<?, ?it/s]
(569, 30) (569,)
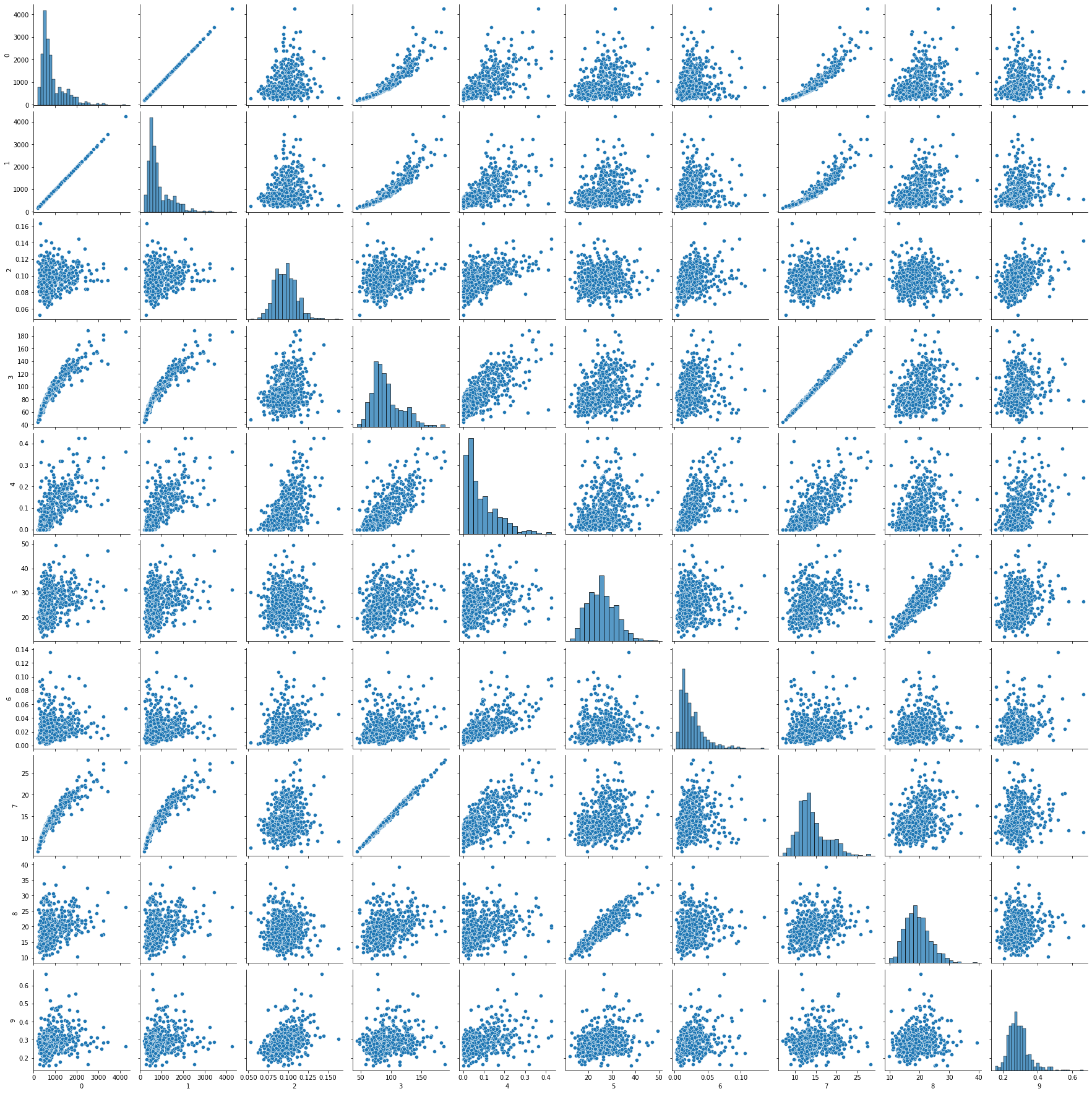
随机选10个特征观察相关性,注意到一些特征间存在很强的线性相关性,此外,很多特征的分布是近似正态分布,比较适合SVM模型分类。
1 2 3 4 5 import matplotlib.pyplot as pltimport seaborn as snsimport pandas as pd0 , 30 , 10 )]))
<seaborn.axisgrid.PairGrid at 0x1a6c91c4d90>
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 from sklearn.model_selection import train_test_splitfrom sklearn.preprocessing import MinMaxScaler, StandardScalerfrom svm import BiLinearSVC, BiKernelSVC0.8 ,42 ,print ("LinearSVM : {:2.4f}%, Kernel SVM : {:2.4f}%" .format (100 , acc2 * 100 ))
LinearSVM : 98.2456%, Kernel SVM : 96.4912%
在Project1中,似乎线性分类器比核RBF效果更好。
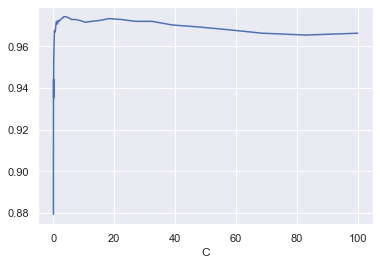
参数选择 线性SVM只有一个超参数$C$,观察不同$C$情况下模型的表现,同时选出最优参数:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 from warnings import filterwarnings'ignore' )2 , 2 , 50 )20 len (C_list), n_iter))for i in tqdm(range (n_iter)):0.8 ,for j, C in enumerate (C_list):2500 ).fit(import seaborn as snsset ()1 )'Test accuracy' )"C" )print ("best C : {}, best accuracy : {:2.4f}%" .format (max () * 100 ,
100%|██████████████████████████████████████████████████████████████████████████████████| 20/20 [00:46<00:00, 2.32s/it]
best C : 3.3932217718953264, best accuracy : 97.4123%
接下来看核SVM的表现,首先用不同的核函数进行实验,发现性能最优的是RBF。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 kernel_list = ["rbf" , "poly" , "sigmoid" ]20 len (kernel_list), n_iter))for i in tqdm(range (n_iter)):0.8 ,for j, kernel in enumerate (kernel_list):5000 ,for i, kernel in enumerate (kernel_list):print ("{:7s}, mean : {:4.2f}%, std : {}" .format (100 ,100 ).std(),
100%|██████████████████████████████████████████████████████████████████████████████████| 20/20 [00:00<00:00, 22.79it/s]
rbf , mean : 96.80%, std : 1.8664844588555454
poly , mean : 94.96%, std : 1.9392711376696599
sigmoid, mean : 96.36%, std : 1.2490377952542226
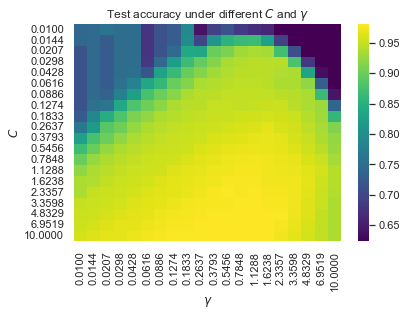
接下来为RBF选取最优参数$C,\gamma$:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 C_list = np.logspace(-2 , 1 , 20 )2 , 1 , 20 )20 len (C_list), len (G_list), n_iter))for i in tqdm(range (n_iter)):0.8 ,for j, C in enumerate (C_list):for k, gamma in enumerate (G_list):2500 ,
100%|██████████████████████████████████████████████████████████████████████████████████| 20/20 [04:21<00:00, 13.06s/it]
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 sns.heatmap(1 ),'viridis' ,"{:.4f}" .format (x) for x in G_list],"{:.4f}" .format (x) for x in C_list],r"$\gamma$" )r"$C$" )r"Test accuracy under different $C$ and $\gamma$" )1 ).argmax()len (G_list), argmax % len (G_list)print ("Best parameters : C={:.4f}, γ={:.4f}, acc={:.2f}%" .format (1 ).flatten()[argmax] * 100 ,
Best parameters : C=6.9519, γ=1.1288, acc=98.11%